Compared Analysis of Retinal Protein Expression Induced by Neutron Radiation and Microgravity
-
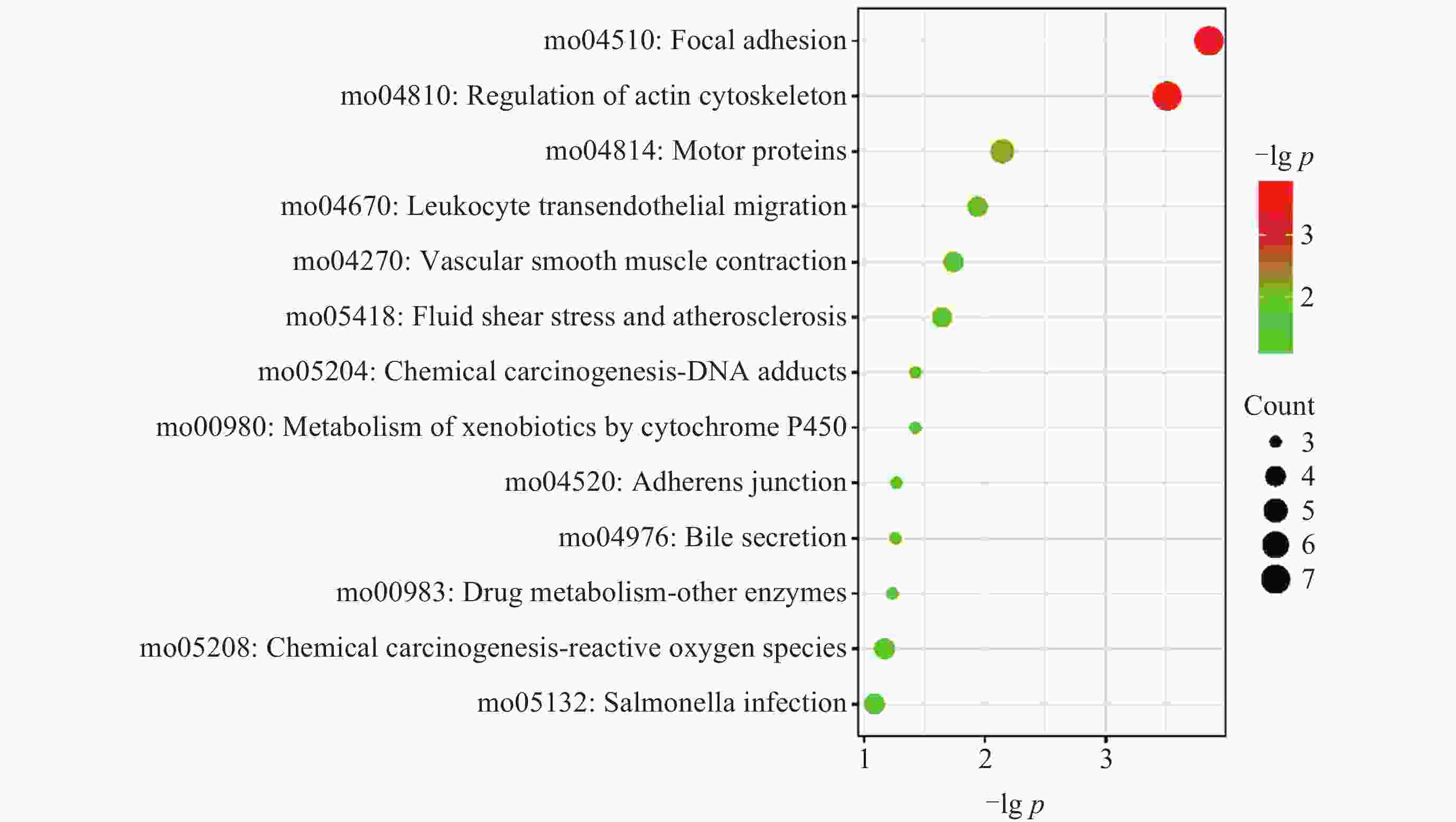
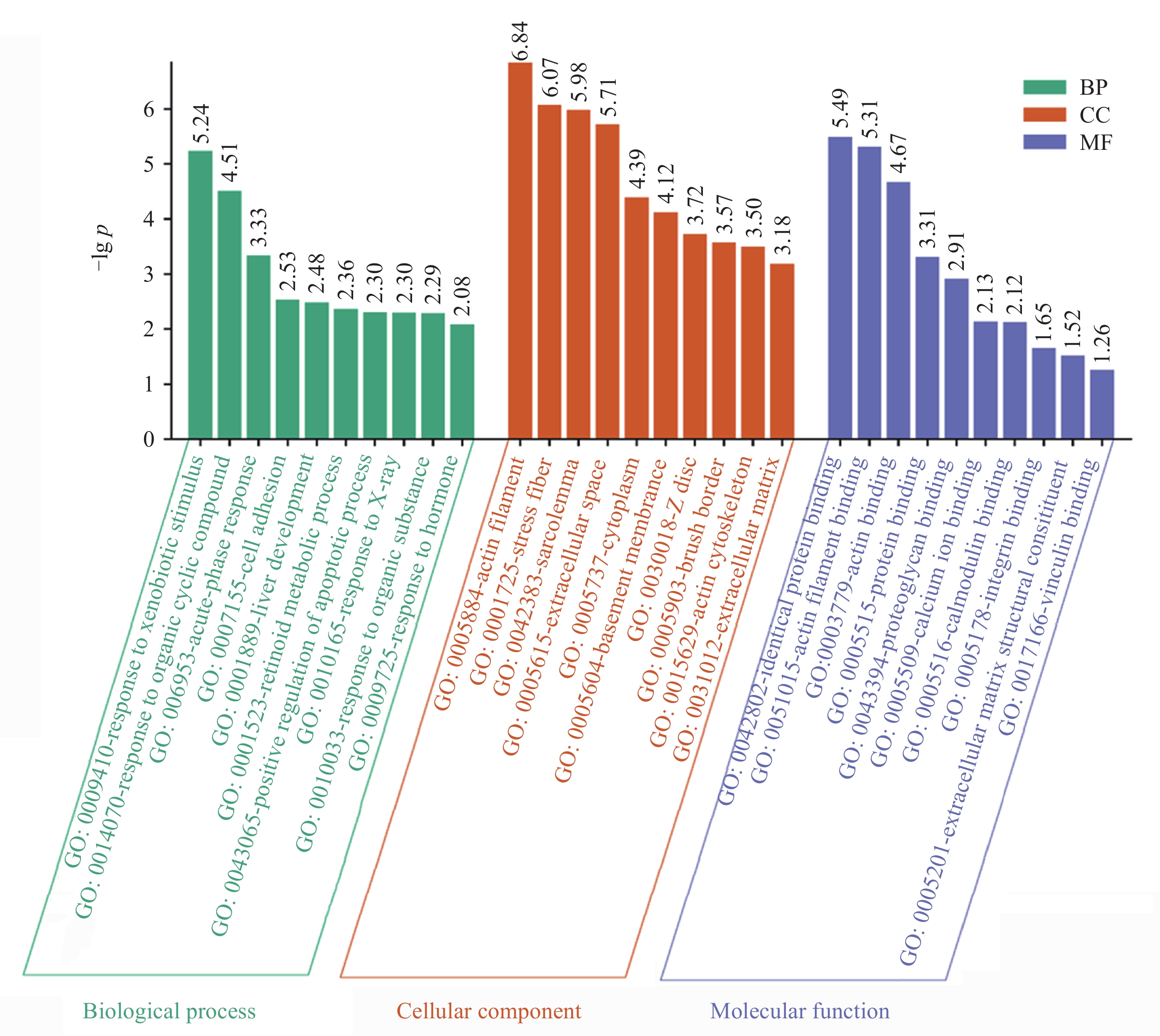
摘要: 研究通过生物信息学方法对比中子辐射与微重力对鼠视网膜蛋白表达的影响, 旨为理解空间环境对视网膜的损伤机制提供生物学基础. 基于差异蛋白数据, 结合基因本体论(GO)、京都基因和基因组百科全书(KEGG)分析及蛋白质-蛋白质相互作用(PPI)网络构建发现: 中子辐射GO分析显著富集于应对外源性刺激, 肌动蛋白丝, 相同蛋白结合, 微重力则主要富集于相机型眼晶状体发育, 线粒体和晶状体结构成分. 中子辐射组KEGG富集于运动蛋白通路, 黏附斑通路和肌动蛋白细胞骨架调控等通路; 微重力组富集于运动蛋白和心肌收缩通路. PPI网络核心模块显示, 中子辐射刺激最显著的生物过程为肌肉收缩, 微重力组最显著的生物过程为相机型眼发育. 在中子辐射和微重力刺激下, 蛋白PMEL, PTN, TPM1, RAB27A表达变化趋势一致, PDPN(Podoplanin)出现相反的变化趋势. 结论揭示视网膜蛋白对中子辐射和微重力刺激的响应存在差异.Abstract: This study employs bioinformatics methods to analyze and compare the effects of neutron radiation and microgravity environments on retinal protein expression in mice, providing a biological basis for understanding the mechanisms of retinal damage induced by space environments. Furthermore, it offers insights for risk assessment and protective measures related to space environments. We obtained differential expression data of retinal proteins in mice exposed to neutron radiation and microgravity environments. Various bioinformatics methods, including GO and KEGG enrichment analyses, PPI network construction and module analysis, and Hub protein screening and analysis, were employed to compare the effects of neutron radiation and microgravity on retinal protein differential expression. The results show that there were differences in the most significantly enriched functions. Neutron radiation inducement primarily enriched in functions such as “response to xenobiotic stimulus”, “actin filament”, and “identical protein binding”. Microgravity inducement enriched in functions such as “lens development in camera-type eye”, “mitochondrion”, and “structural constituent of eye lens”. The KEGG analysis showed that the changes in the “Motor proteins” pathway were consistent under both neutron radiation and microgravity inducement. In addition, neutron radiation inducement also enriched in the “Focal adhesion” and “Regulation of actin cytoskeleton” pathways, while microgravity inducement also enriched in the “Cardiac muscle contraction” pathway. The Hub proteins and Biological Process (BP) of the most significant modules were different. The most significant BP under neutron radiation inducement was “muscle contraction”, while under microgravity inducement, “camera-type eye development” was the most significant. Under different inducement, the expression trends of proteins PMEL, PTN, TPM1, and RAB27A were consistent. However, PDPN showed an opposite change. The results suggest that retinal proteins respond differently to neutron radiation and microgravity stimuli. These findings have the potential to elucidate the mechanisms of retinal damage caused by space radiation or microgravity and provide a reference for developing targeted protective measures.
-
Key words:
- Neutron radiation /
- Microgravity /
- Retina /
- Differentially expressed protein
-
表 1 中子辐射诱导视网膜蛋白差异表达枢纽蛋白的节点度(前10个)
Table 1. Node degree of differentially expressed Hub proteins in the retina induced by neutron radiation (top 10)
Rank Name Score 1 VCL 14 2 MYH11 9 3 MYL9 8 3 TAGLN 8 3 MYLK 8 3 TPM2 8 3 TPM1 8 8 CALD1 7 8 FLNA 7 10 ANXA5 6 表 2 最重要模块的蛋白组成(中子辐射)
Table 2. Protein composition of the most important module (neutron radiation)
Name Log2FC p-value VCL 0.315672606 0.003270326 TAGLN 1.306693581 0.000112783 MYLK 0.842655794 0.001317789 MYL9 1.258456494 0.000221689 TPM1 0.792268363 0.000407139 TPM2 1.284591944 0.000206282 CALD1 0.509000461 0.000924537 MYH11 1.468484826 5.69×10–5 表 3 最重要模块的部分功能(中子辐射)
Table 3. Partial functions of the most important module (neutron radiation)
GO-ID p-value Description Genes in test set 6936 3.12×10–15 Muscle contraction CALD1, TPM2, TPM1, MYH11, MYL9, VCL, MYLK 3012 8.59×10–15 Muscle system process CALD1, TPM2, TPM1, MYH11, MYL9, VCL, MYLK 3008 9.35×10–6 System process CALD1, TPM2, TPM1, MYH11, MYL9, VCL, MYLK 43462 2.13×10–6 Regulation of ATPase activity TPM2, TPM1 32501 6.22×10–5 Multicellular organismal process TAGLN, CALD1, TPM2, TPM1, MYH11, MYL9, VCL, MYLK 30811 1.99×10–3 Regulation of nucleotide catabolic process TPM2, TPM1 33121 1.99×10–3 Regulation of purine nucleotide catabolic process TPM2, TPM1 48251 2.24×10–3 Elastic fiber assembly MYH11 43297 2.80×10–3 Apical junction assembly VCL 7517 2.90×10–3 Muscle organ development TAGLN, MYH11 表 4 微重力环境诱导视网膜蛋白差异表达枢纽蛋白的节点度(前10个)
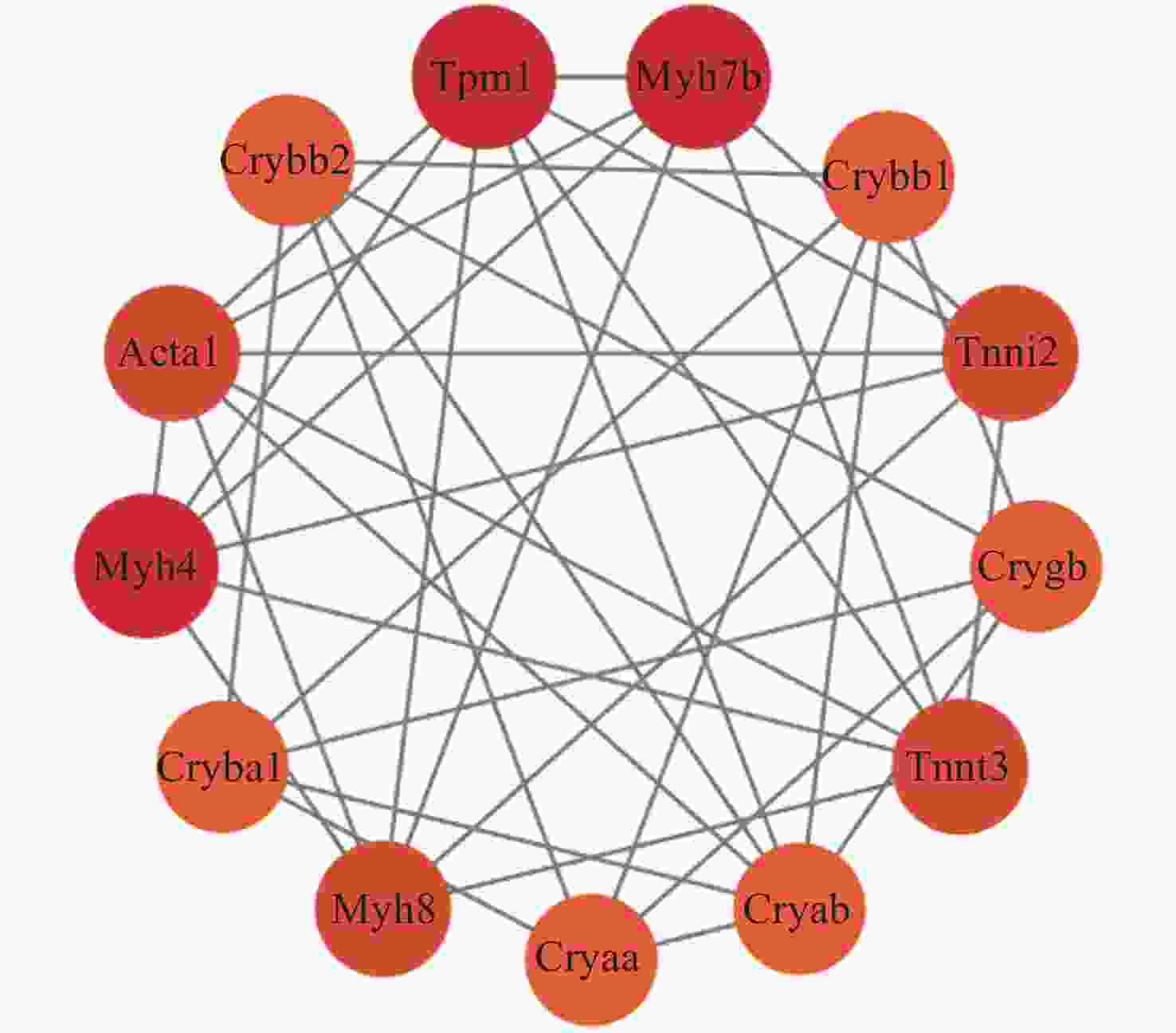
Table 4. Node degree of hub proteins differentially expressed in the retina induced by microgravity environment (top 10)
Name Rank Score ACTA1 1 13 CRYAB 2 12 TPM1 3 11 TNNT3 4 9 TNNI2 4 9 MYH4 6 8 MYH8 6 8 CSRP3 6 8 CRYBB1 9 7 PXN 9 7 表 5 最重要模块的蛋白组成(微重力环境)
Table 5. Protein composition of the most important module (microgravity environment)
Name Log2FC FDR adjusted MYH8 –2.680179586 0.008307896 CRYBA1 –4.504364715 0.048282442 MYH4 –3.854188267 0.03172948 ACTA1 –4.332887144 0.03201434 CRYBB2 –3.667393599 0.047707924 TPM1 4.416731705 0.00830411 MYH7 B –1.197124303 0.020782533 CRYBB1 –4.618506708 0.048847149 TNNI2 –3.456403098 0.047905536 CRYGB –4.481399359 0.048586694 TNNT3 –4.306619987 0.031979082 CRYAB –3.475877689 0.048783752 CRYAA –3.132446621 0.048329001 表 6 最重要模块的部分功能(微重力环境)
Table 6. Partial functions of the most important module (microgravity environment)
GO-ID p-value Description Genes in test set 43010 9.94×10–10 Camera-type eye development CRYGB, CRYBA1, CRYBB2, CRYAA, CRYAB 1654 2.54×10–9 Eye development CRYGB, CRYBA1, CRYBB2, CRYAA, CRYAB 7423 3.05×10–8 Sensory organ development CRYGB, CRYBA1, CRYBB2, CRYAA, CRYAB 2088 3.15×10–7 Lens development in camera-type eye CRYGB, CRYAA, CRYAB 60561 3.93×10–7 Apoptosis involved in morphogenesis CRYAA, CRYAB 70307 3.93×10–7 Lens fiber cell development CRYGB, CRYAA 70309 3.93×10–7 Lens fiber cell morphogenesis CRYGB, CRYAA 48513 5.86×10–7 Organ development ACTA1, CRYGB, TPM1, CRYBA1, CRYBB2, CRYAA, CRYAB 7021 7.87×10–7 Tubulin complex assembly CRYAA, CRYAB 48731 3.12×10–6 System development ACTA1, CRYGB, TPM1, CRYBA1, CRYBB2, CRYAA, CRYAB 表 7 中子辐射诱导视网膜PMEL, PTN, TPM1, RAB27A, PDPN差异表达情况
Table 7. Differential expression of PMEL, PTN, TPM1, RAB27A, and PDPN in the retina induced by neutron radiation
Name Log2FC p-value Expression PMEL 1.517216215 0.004936262 Up PTN 0.538650012 0.015295976 Up TPM1 0.792268363 0.000407139 Up RAB27A 0.559708733 0.01240258 Up PDPN –0.390614025 0.015486769 Down 表 8 微重力环境诱导视网膜PMEL, PTN, TPM1, RAB27A, PDPN差异表达情况
Table 8. Differential expression of PMEL, PTN, TPM1, RAB27A, and PDPN in the retina induced by microgravity environment
Name Log2FC FDR adjusted Expression PMEL 5.279723053 0.00832942 Up PTN 5.278221595 0.008310423 Up TPM1 4.416731705 0.00830411 Up RAB27A 2.720412802 0.031670154 Up PDPN 2.148468927 0.048066344 Up -
[1] MAO X W, BOERMA M, RODRIGUEZ D, et al. Acute effect of low-dose space radiation on mouse retina and retinal endothelial cells[J]. Radiation Research, 2018, 190(1): 45-52 doi: 10.1667/RR14977.1 [2] VAN REYK D M, GILLIES M C, DAVIES M J. The retina: oxidative stress and diabetes[J]. Redox Report, 2003, 8(4): 187-192 doi: 10.1179/135100003225002673 [3] LEE A G, MADER T H, GIBSON C R, et al. Spaceflight Associated Neuro-ocular Syndrome (SANS) and the neuro-ophthalmologic effects of microgravity: a review and an update[J]. NPJ Microgravity, 2020, 6(1): 7 doi: 10.1038/s41526-020-0097-9 [4] MADER T H, GIBSON C R, PASS A F, et al. Optic disc edema, globe flattening, choroidal folds, and hyperopic shifts observed in astronauts after long-duration space flight[J]. Ophthalmology, 2011, 118(10): 2058-2069 doi: 10.1016/j.ophtha.2011.06.021 [5] VYAS R J, YOUNG M, MURRAY M C, et al. Decreased vascular patterning in the retinas of astronaut crew members as new measure of ocular damage in spaceflight-associated neuro-ocular syndrome[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2020, 61(14): 34 [6] MAO X W, BYRUM S, NISHIYAMA N C, et al. Impact of spaceflight and artificial gravity on the mouse retina: biochemical and proteomic analysis[J]. International Journal of Molecular Sciences, 2018, 19(9): 2546 doi: 10.3390/ijms19092546 [7] FENG J D, ZHAO X D, LUO Y Z, et al. Effects of neutron irradiation on ophthalmic fundus structure, visual function and the mechanisms underlying these effects in rats[J]. Acta Astronautica, 2021, 186: 403-417 doi: 10.1016/j.actaastro.2021.04.032 [8] MAO X W, NISHIYAMA N C, BYRUM S D, et al. Characterization of mouse ocular response to a 35-day spaceflight mission: evidence of blood-retinal barrier disruption and ocular adaptations[J]. Scientific Reports, 2019, 9(1): 8215 doi: 10.1038/s41598-019-44696-0 [9] SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Research, 2003, 13(11): 2498-2504 doi: 10.1101/gr.1239303 [10] BADER G D, HOGUE C W V. An automated method for finding molecular complexes in large protein interaction networks[J]. BMC Bioinformatics, 2003, 4(1): 2 doi: 10.1186/1471-2105-4-2 [11] MAERE S, HEYMANS K, KUIPER M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks[J]. Bioinformatics, 2005, 21(16): 3448-3449 doi: 10.1093/bioinformatics/bti551 [12] JEONG H, TOMBOR B, ALBERT R, et al. The large-scale organization of metabolic networks[J]. Nature, 2000, 407(6804): 651-654 doi: 10.1038/35036627 [13] CHIN C H, CHEN S H, WU H H, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome[J]. BMC Systems Biology, 2014, 8(4): S11 [14] MU Y X, ZHANG N, WEI D Y, et al. Müller cells are activated in response to retinal outer nuclear layer degeneration in rats subjected to simulated weightlessness conditions[J]. Neural Regeneration Research, 2025, 20(7): 2116-2128 doi: 10.4103/NRR.NRR-D-23-01035 [15] MIYARA N, SHINZATO M, YAMASHIRO Y, et al. Proteomic analysis of rat retina in a steroid-induced ocular hypertension model: potential vulnerability to oxidative stress[J]. Japanese Journal of Ophthalmology, 2008, 52(2): 84-90 doi: 10.1007/s10384-007-0507-5 [16] ASHBY R S, MEGAW P L, MORGAN I G. Changes in retinal αB-crystallin (cryab) RNA transcript levels during periods of altered ocular growth in chickens[J]. Experimental Eye Research, 2010, 90(2): 238-243 doi: 10.1016/j.exer.2009.10.011 [17] ZHANG S B, LIU C P, WANG Q, et al. CRYAA and GJA8 promote visual development after whisker tactile deprivation[J]. Heliyon, 2023, 9(3): e13897 doi: 10.1016/j.heliyon.2023.e13897 [18] XU Q, BAI Y J HUANG L Z, et al. Knockout of αA-crystallin inhibits ocular neovascularization[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2015, 56(2): 816-826 [19] FORT P E, LAMPI K J. New focus on alpha-crystallins in retinal neurodegenerative diseases[J]. Experimental Eye Research, 2011, 92(2): 98-103 doi: 10.1016/j.exer.2010.11.008 [20] BOYA P, KAARNIRANTA K, HANDA J T, et al. Lysosomes in retinal health and disease[J]. Trends in Neurosciences, 2023, 46(12): 1067-1082 doi: 10.1016/j.tins.2023.09.006 [21] VALAPALA M, EDWARDS M, HOSE S, et al. βA3/A1-crystallin is a critical mediator of STAT3 signaling in optic nerve astrocytes[J]. Scientific Reports, 2015, 5(1): 8755 doi: 10.1038/srep08755 [22] ZIGLER JR J S, SINHA D. βA3/A1-crystallin: more than a lens protein[J]. Progress in Retinal and Eye Research, 2015, 44: 62-85 doi: 10.1016/j.preteyeres.2014.11.002 [23] BÖHM M R R, PFROMMER S, CHIWITT C, et al. Crystallin-β-b2-overexpressing NPCs support the survival of injured retinal ganglion cells and photoreceptors in rats[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2012, 53(13): 8265-8279 [24] LIEDTKE T, SCHWAMBORN J C, SCHRÖER U, et al. Elongation of axons during regeneration involves retinal crystallin βb2 (crybb2)[J]. Molecular :Times New Roman;">& Cellular Proteomics, 2007, 6(5): 895-907 [25] LIU H H, BELL K, HERRMANN A, et al. Crystallins play a crucial role in glaucoma and promote neuronal cell survival in an in vitro model through modulating Müller cell secretion[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2022, 63(8): 3 [26] HODGES E D, CHRYSTAL P W, FOOTZ T, et al. Disrupting the repeat domain of premelanosome protein (PMEL) produces dysamyloidosis and dystrophic ocular pigment reflective of pigmentary glaucoma[J]. International Journal of Molecular Sciences, 2023, 24(19): 14423 doi: 10.3390/ijms241914423 [27] ZHU X, BAI Y, YU W, et al. The effects of pleiotrophin in proliferative diabetic retinopathy[J]. PloS One, 2015, 10(1): e0115523 doi: 10.1371/journal.pone.0115523 [28] LAMPROU M, KASTANA P, KOFINA F, et al. Pleiotrophin selectively binds to vascular endothelial growth factor receptor 2 and inhibits or stimulates cell migration depending on αν β3 integrin expression[J]. Angiogenesis, 2020, 23(4): 621-636 doi: 10.1007/s10456-020-09733-x [29] LI R, LIANG Y X, LIN B. Accumulation of systematic TPM1 mediates inflammation and neuronal remodeling by phosphorylating PKA and regulating the FABP5/NF‐κB signaling pathway in the retina of aged mice[J]. Aging Cell, 2022, 21(3): e13566 doi: 10.1111/acel.13566 [30] RONG L, JING Z, MENG C, et al. TPM1 mediates inflammation by regulating PKA/CREB signaling pathway[J]. Investigative Ophthalmology :Times New Roman;">& Visual Science, 2022, 63(7): 4125-F0362 [31] IACONO D, HATCH K, MURPHY E K, et al. Proteomic changes in the hippocampus of large mammals after total-body low dose radiation[J]. PloS One, 2024, 19(3): e0296903 doi: 10.1371/journal.pone.0296903 [32] KLOMP A E, TEOFILO K, LEGACKI E, et al. Analysis of the linkage of MYRIP and MYO7A to melanosomes by RAB27A in retinal pigment epithelial cells[J]. Cell Motility, 2007, 64(6): 474-487 doi: 10.1002/cm.20198 [33] REICHHART N, MARKOWSKI M, ISHIYAMA S, et al. Rab27a GTPase modulates L-type Ca2+ channel function via interaction with the II–III linker of CaV1.3 subunit[J]. Cellular Signalling, 2015, 27(11): 2231-2240 doi: 10.1016/j.cellsig.2015.07.023 [34] OUCHI T, KONO K, SATOU R, et al. Upregulation of Amy1 in the salivary glands of mice exposed to a lunar gravity environment using the multiple artificial gravity research system[J]. Frontiers in Physiology, 2024, 15: 1417719 doi: 10.3389/fphys.2024.1417719 [35] RETZBACH E P, SHEEHAN S A, KRISHNAN H, et al. Independent effects of Src kinase and podoplanin on anchorage independent cell growth and migration[J]. Molecular Carcinogenesis, 2022, 61(7): 677-689 doi: 10.1002/mc.23410 [36] RUSU M C, NICOLESCU M I, VRAPCIU A D. Evidence of lymphatics in the rat eye retina[J]. Annals of Anatomy-Anatomischer Anzeiger, 2022, 244: 151987 doi: 10.1016/j.aanat.2022.151987 [37] ESHAQ R, WARAR R, HARRIS N. Upregulation of Prox‐1, podoplanin and LYVE‐1 in retinal endothelial cells under hyperglycemic conditions[J]. The FASEB Journal, 2020, 34(S1): 1 [38] BONENTE D, BIANCHI L, DE SALVO R, et al. Co-Expression of podoplanin and CD44 in proliferative vitreoretinopathy epiretinal membranes[J]. International Journal of Molecular Sciences, 2023, 24(11): 9728 doi: 10.3390/ijms24119728 [39] BADHWAR G D, KEITH J E, CLEGHORN T F. Neutron measurements onboard the space shuttle[J]. Radiation Measurements, 2001, 33(3): 235-241 doi: 10.1016/S1350-4487(00)00159-1 [40] ZEITLIN C, CASTRO A J, BEARD K B, et al. Results from the radiation assessment detector on the international space station: Part 1, the charged particle detector[J]. Life Sciences in Space Research, 2023, 39: 67-75 doi: 10.1016/j.lssr.2023.01.003 [41] PROSS H D, KOST M, KIEFER J. Repair of radiation induced genetic damage under microgravity[J]. Advances in Space Research, 1994, 14(10): 125-130 doi: 10.1016/0273-1177(94)90461-8 [42] HORNECK G, RETTBERG P, KOZUBEK S, et al. The influence of microgravity on repair of radiation-induced DNA damage in bacteria and human fibroblasts[J]. Radiation Research, 1997, 147(3): 376-384 doi: 10.2307/3579347 [43] STRIKE T A. Acute mortality of mice and rats exposed to 14 MeV neutrons[J]. Radiation Research, 1970, 43(3): 679-690 doi: 10.2307/3573237 [44] NILSSON S, HELOU K, WALENTINSSON A, et al. Rat–mouse and rat–human comparative maps based on gene homology and high-resolution zoo-FISH[J]. Genomics, 2001, 74(3): 287-298 doi: 10.1006/geno.2001.6550 -
-





 孙思敏 女, 2002年1月出生于河北省邯郸市, 现为南京航空航天大学能源动力专业研究生, 主要研究方向为辐射生物效应与辐射风险评估等. E-mail:
孙思敏 女, 2002年1月出生于河北省邯郸市, 现为南京航空航天大学能源动力专业研究生, 主要研究方向为辐射生物效应与辐射风险评估等. E-mail: 

 下载:
下载: